Data Analysis using Sparks, Pandas, and Matplotlib using Jupyter Notebook for data in S3(Minio)
By Prashant Shahi
- 13 minutes read - 2709 wordsData Analysis is to understand problems facing an organization and to explore data in meaningful ways. Data in itself is merely facts and figures. Evaluation of the data can provide advantages to the organization and aid in making business decisions.
Brief Overview of the components
Apache Spark is a lightning-fast cluster computing technology, designed for fast computation and based on Hadoop MapReduce.
Pandas is a software library written in Python for data manipulation and analysis. Similarly, Matplotlib is another python library used for 2D plotting which produces publication quality figures in a variety of hardcopy formats and interactive environments across platforms.
Jupyter Notebook is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations, and narrative text.
Minio is an open-source cloud object storage server which follows Amazon S3 protocol and at times, referred to as an Open-Source Amazon S3 alternative, which is available for anyone on internet for free to deploy on their machines.
Setup
System configuration (VM/Instance)
The system configuration selected for the task is as mentioned below :
- 6 cores CPUs
- 10 GB memory (RAM)
- 200 GB Disk Size (ROM)
It should be fine even if your machine configurations are lower the one used here.
Downloading Minio Server and Client
Minio Server
Follow the steps below to setup Minio Server :
# Downloading Minio binary and copying to /opt
sudo wget -O /opt/minio https://dl.minio.io/server/minio/release/linux-amd64/minio
# Changing the file permission of binary to mark as executable
sudo chmod +x /opt/minio
# Creating a symbolic link to /usr/local/bin to make the file executable from any path
sudo ln -s /opt/minio /usr/local/bin/
# Making the data directory for storing the objects/data for minio server
mkdir ./data
# Running the Minio server with the data directory parameter
minio server ./data
Minio Client
Follow the steps below to setup Minio Client :
# Downloading minio binary and copying to /opt
sudo wget -O /opt/mc https://dl.minio.io/client/mc/release/linux-amd64/mc
# Changing the file permission of binary to mark as executable
sudo chmod +x /opt/mc
# Creatin a symbolic link to /usr/local/bin to make the file executable from any path
sudo ln -s /opt/mc /usr/local/bin
# Executing the command with help parameter to ensure that installation was a success
mc --help
Loading Sample Data
Follow the steps below how to load a Sample Data to S3 using Minio Client :
# Downloading the sample data of TotalPopulationBySex.csv from UN
wget -O TotalPopulation.csv "https://esa.un.org/unpd/wpp/DVD/Files/1_Indicators%20(Standard)/CSV_FILES/WPP2017_TotalPopulationBySex.csv"
# Compressing the csv file
gzip TotalPopulation.csv
# Creating a new bucket
mc mb data/mycsvbucket
# Copying the compressed file inside bucket
mc cp TotalPopulation.csv.gz data/mycsvbucket/
Setting up Java Environment for Spark Shell
# Adding ppa to local repository
sudo add-apt-repository ppa:webupd8team/java
# Updating repository archives
sudo apt update
# Installing Oracle Java8
sudo apt install -y oracle-java8-installer
# Verifying the java installation
javac -version
# Setting Oracle Java8 as default (In case of multiple java versions)
sudo apt install -y oracle-java8-set-default
# Setting up environment variable (Also, add this to the `~/.bashrc` file to apply for next boot)
export JAVA_HOME=/usr/lib/jvm/java-8-oracle
export PATH=$PATH:$JAVA_HOME/bin
Installation of Apache Spark and Hadoop
Steps to install Apache Spark is as follow :
# Download Spark v2.3.0 without Hadoop
wget http://archive.apache.org/dist/spark/spark-2.3.0/spark-2.3.0-bin-without-hadoop.tgz
# Extracting the compressed file
sudo tar -C /opt/ -xvf spark-2.3.0-bin-without-hadoop.tgz
# Setting up environment variable (Also, add this to the `~/.bashrc` file to apply for next boot)
export SPARK_HOME=/opt/spark-2.3.0-bin-without-hadoop
export PATH=$PATH:$SPARK_HOME/bin
Steps to install Apache Hadoop is as follow :
# Download Hadoop v2.8.2
wget https://archive.apache.org/dist/hadoop/core/hadoop-2.8.2/hadoop-2.8.2.tar.gz
# Extracting the compressed file
sudo tar -C /opt/ -xvf hadoop-2.8.2.tar.gz
# Setting up environment for Hadoop
export HADOOP_HOME=/opt/hadoop-2.8.2
export PATH=$PATH:$HADOOP_HOME/bin
export SPARK_DIST_CLASSPATH=$(hadoop classpath)
export LD_LIBRARY_PATH=$HADOOP_HOME/lib/native
Setting up Minio Server endpoint and credentials
Open the file $HADOOP_HOME/etc/hadoop/core-site.xml for editing. In the example XML file below, Minio server is running at http://127.0.0.1:9000 with access key minio and secret key minio123. Make sure to update relevant sections with valid Minio server endpoint and credentials.
<?xml version="1.0" encoding="UTF-8"?>
<?xml-stylesheet type="text/xsl" href="configuration.xsl"?>
<configuration>
<property>
<name>fs.s3a.endpoint</name>
<description>AWS S3 endpoint to connect to. An up-to-date list is
provided in the AWS Documentation: regions and endpoints. Without this
property, the standard region (s3.amazonaws.com) is assumed.
</description>
<value>http://127.0.0.1:9000</value>
</property>
<property>
<name>fs.s3a.access.key</name>
<description>AWS access key ID.</description>
<value>minio</value>
</property>
<property>
<name>fs.s3a.secret.key</name>
<description>AWS secret key.</description>
<value>minio123</value>
</property>
<property>
<name>fs.s3a.path.style.access</name>
<value>true</value>
<description>Enable S3 path style access ie disabling the default virtual hosting behaviour.
Useful for S3A-compliant storage providers as it removes the need to set up DNS for virtual hosting.
</description>
</property>
<property>
<name>fs.s3a.impl</name>
<value>org.apache.hadoop.fs.s3a.S3AFileSystem</value>
<description>The implementation class of the S3A Filesystem</description>
</property>
</configuration>
Get started
Spark Shell on CSV in Minio (S3)
Note: Make sure JAVA_HOME has been set before setting up Spark Shell.
Spark-Select can be integrated with Spark via spark-shell, pyspark, spark-submit, etc. You can also add it as Maven dependency, sbt-spark-package or a jar import.
Let’s go through the steps below to use spark-shell in an example.
-
Start Minio server and configure mc to interact with this server.
-
Create a bucket and upload a sample file :
# Downloading sample csv
wget "https://gist.github.com/raw/dc0d42feb6e8c816ed2ff1778b35a130/people.csv"
# Creating a bucket named sjm-airlines
mc mb data/sjm-airlines
# Copying the csv to the created bucket using minio client
mc cp people.csv data/sjm-airlines
- Download the sample scala code:
wget "https://gist.github.com/raw/dc0d42feb6e8c816ed2ff1778b35a130/csv.scala"
- Downloading depedencies and adding it to spark :
# Creating jars folder
mkdir jars
# Changing current directory to ./jars
cd jars
# Downloading jar dependencies
wget http://central.maven.org/maven2/org/apache/hadoop/hadoop-aws/2.8.2/hadoop-aws-2.8.2.jar
wget http://central.maven.org/maven2/org/apache/httpcomponents/httpclient/4.5.3/httpclient-4.5.3.jar
wget http://central.maven.org/maven2/joda-time/joda-time/2.9.9/joda-time-2.9.9.jar
wget http://central.maven.org/maven2/com/amazonaws/aws-java-sdk-s3/1.11.524/aws-java-sdk-s3-1.11.524.jar
wget http://central.maven.org/maven2/com/amazonaws/aws-java-sdk-core/1.11.524/aws-java-sdk-core-1.11.524.jar
wget http://central.maven.org/maven2/com/amazonaws/aws-java-sdk/1.11.524/aws-java-sdk-1.11.524.jar
wget http://central.maven.org/maven2/com/amazonaws/aws-java-sdk-kms/1.11.524/aws-java-sdk-kms-1.11.524.jar
# Copying all the jars to $SPARK_HOME/jars/
cp *.jar $SPARK_HOME/jars/
-
Configure Apache Spark with Minio. Detailed steps are available in this document.
-
Let’s start
spark-shellwith the following command. To load some additional package later on, you can use--packagesflag as well.
$SPARK_HOME/bin/spark-shell --master local[4]
- After
spark-shellis successfully invoked, load the csv.scala, and display the data:
Welcome to
____ __
/ __/__ ___ _____/ /__
_\ \/ _ \/ _ `/ __/ '_/
/___/ .__/\_,_/_/ /_/\_\ version 2.3.0
/_/
Using Scala version 2.XX.XX (Java HotSpot(TM) 64-Bit Server VM, Java 1.8.0_XXX)
Type in expressions to have them evaluated.
Type :help for more information.
scala> :load csv.scala
Loading examples/csv.scala...
import org.apache.spark.sql._
import org.apache.spark.sql.types._
defined object app
scala> app.main(Array())
+-------+---+
| name|age|
+-------+---+
|Michael| 31|
| Andy| 30|
| Justin| 19|
+-------+---+
+-------+---+
| name|age|
+-------+---+
|Michael| 31|
| Andy| 30|
+-------+---+
scala>
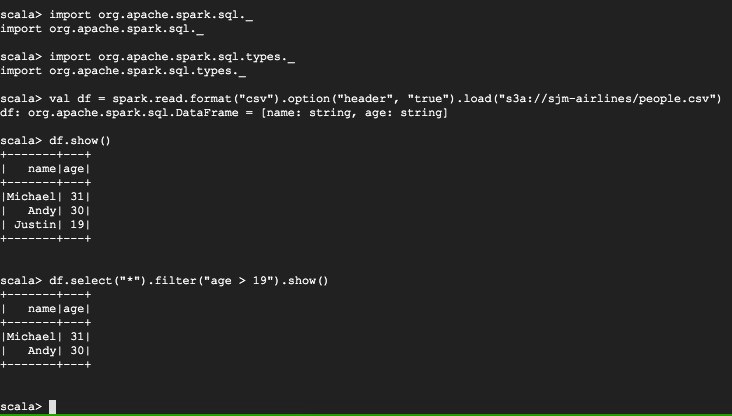
You can see that out of 3 entries, we could use SQL-like query to only select those entries with age > 19.
Awesome, you have successfully set up Spark! Let’s proceed futher.
Spark-Shell using PySpark and Minio
Make sure all of the aws-java-sdk jars are present under $SPARK_HOME/jars/ or
added to the spark.jars.packages in spark-defaults.conf file, before proceeding.
# Running pyspark from Spark_Home Binary
$SPARK_HOME/bin/pyspark
``
You should see the following screen :
```python
Welcome to
____ __
/ __/__ ___ _____/ /__
_\ \/ _ \/ _ `/ __/ '_/
/__ / .__/\_,_/_/ /_/\_\ version 2.3.0
/_/
Using Python version 2.7.12 (default, Nov 12 2018 14:36:49)
SparkSession available as 'spark'.
>>>
Let’s execute the commands following lines to verify the same as in scala-shell can be achieved in PySpark:
>>> from pyspark.sql.types import *
>>> df = spark.read.format("csv").option("header", "true").load("s3a://sjm-airlines/people.csv")
>>> df.show()
+-------+---+
| name|age|
+-------+---+
|Michael| 31|
| Andy| 30|
| Justin| 19|
+-------+---+
>>> df.select("*").filter("age > 19").show()
+-------+---+
| name|age|
+-------+---+
|Michael| 31|
| Andy| 30|
+-------+---+
Connect Minio and Spark with Jupyter Notebook
Follow the steps below to set up Jupyter:
# Downloading shell script to install Jupyter using Anaconda
wget https://repo.anaconda.com/archive/Anaconda3-2018.12-Linux-x86_64.sh
# Making the shell script executable
chmod +x ./Anaconda3-2018.12-Linux-x86_64.sh
# Running the shell script with bash
bash Anaconda3-2018.12-Linux-x86_64.sh
# Create new conda environment with minimal environment with only python installed.
conda create -n myconda python=3
# Put your self inside this environment run
conda activate myconda
# Verify the Anaconda Python v3.x Terminal inside the environment. To exit, press CTRL+D or `exit()`
python
# Install Jupyter Notebook inside the environment
conda install jupyter
# Install findspark inside the environment using conda-forge channel
conda install -c conda-forge findspark
# (Optional) Setting up jupyter notebook password, enter the desired password (If not set, have to use randomly generated tokens each time)
jupyter notebook password
# Running Jupyter Notebook and making it available to public at port 8888
jupyter notebook --ip 0.0.0.0 --port 8888
You should be seeing the following, if everything goes well :
[I 06:50:01.156 NotebookApp] JupyterLab extension loaded from /home/prashant/anaconda3/lib/python3.7/site-packages/jupyterlab
[I 06:50:01.157 NotebookApp] JupyterLab application directory is /home/prashant/anaconda3/share/jupyter/lab
[I 06:50:01.158 NotebookApp] Serving notebooks from local directory: /home/prashant
[I 06:50:01.158 NotebookApp] The Jupyter Notebook is running at:
[I 06:50:01.158 NotebookApp] http://(instance-1 or 127.0.0.1):8888/
[I 06:50:01.158 NotebookApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
Deactivate Conda virtual environment
Example:
# Usage: conda deactivate <environment-name>
conda deactivate myconda
Converting python scripts(.py) file to Jupyter notebook(.ipynb) file
# Installing p2j using python-pip
pip install p2j
Example:
# Generating .ipynb file out of some sample script.py using p2j
p2j script.py
Converting Jupiter notebook(.ipynb) file to python scripts(.py) file
You can make use of nbconvert that comes along with Jupiter. Example:
# Generating script.py file out of some sample .ipynb file using jupyter nbconvert
jupyter nbconvert script.ipynb
Creating a sample python file
Let’s create a python file spark-minio.py with the codes below :
# Import sys and print the python environment
import sys
print(sys.executable)
# Import findspark to find spark make it accessible at run time
import findspark
findspark.init()
# Import pyspark and its components
import pyspark
from pyspark.sql.types import *
from pyspark.sql import SparkSession
# Creating SparkSession
spark = SparkSession.builder.getOrCreate()
# Creating schema of the CSV fields
schema = StructType([StructField('name', StringType(), True),StructField('age', IntegerType(), True)])
# Creating a dataframe from a csv in S3
df = spark.read.format("csv").option("header", "true").load("s3a://sjm-airlines/people.csv")
# Displaying all data in the CSV
df.show()
# Displaying all the data in the csv for which age is greater than 19
df.select("*").filter("age > 19").show()
Now, converting the python code(spark-minio.py) to jupyter notebook compatible file (.ipynb) :
# Generating spark-minio.ipynb file out of spark-minio.py
p2j spark-minio.py
Running .ipynb file from the Jupyter notebook UI
Let’s open the UI running at http://(server-public-ip-address/localhost):8888/. Enter the jupyter notebok password (or the token) and then, you should be seeing something like this :
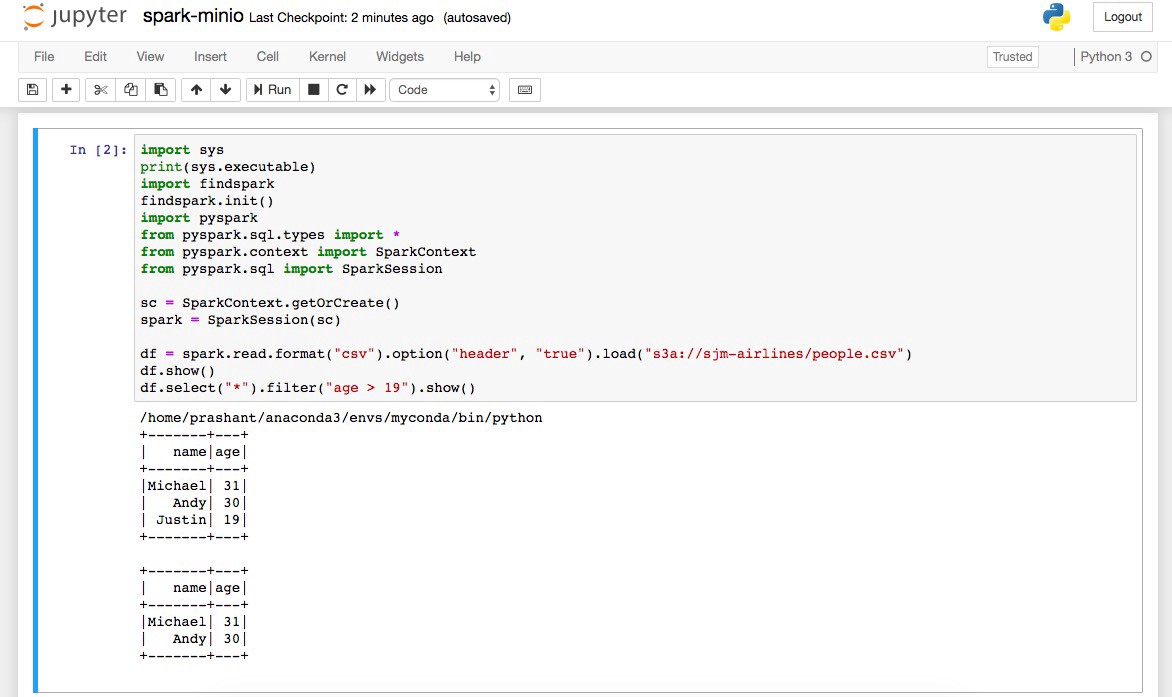
Select spark-minio.ipynb file and click on run, if everything went right, you should be getting the screen below :
Running Some Live Examples Before running the example, let’s get compress the sample csv file with gzip compression.
# Generating gzip file named people.csv.gz file out of people.csv, while keeping the original file with -k flag
gzip -k people.csv
# Copying the csv.gz file to the bucket using minio client
mc cp people.csv.gz data/sjm-airlines
In Jupyter Notebook, go to File Tab > New Notebook > Python 3 (Or any other kernel). Try the following pyspark example on the data present in Minio. Note that the gzip compression is automatically detected with the .gz extension and handled when loading it with Spark’s native csv format.
import findspark
findspark.init()
import pyspark
from pyspark.sql.types import *
from pyspark.sql import SparkSession
spark = SparkSession.builder.getOrCreate()
schema = StructType([StructField('name', StringType(), True),StructField('age', IntegerType(), True)])
df = spark.read.format("csv").option("header", "true").load("s3a://sjm-airlines/people.csv.gz")
df.createOrReplaceTempView("people")
print("List of all people :")
df2.show()
print("People with age less than 20 :")
df2 = spark.sql("SELECT * FROM people where age>20")
df2.show()
If the steps are properly followed, you should be seeing the following in the jupyter notebook:
List of all people :
+-------+---+
| name|age|
+-------+---+
|Michael| 31|
| Andy| 30|
+-------+---+
People with age less than 20 :
+-------+---+
| name|age|
+-------+---+
|Michael| 31|
| Andy| 30|
+-------+---+
For the next example, we are gonna use SQL query capability of Spark dataframe on comparatively big CSV with 13 header fields and 2000251 entries. For the task, at first, we are gonna download the CSV with gzipped compression from the following link.
wget https://gist.github.com/raw/d65254b1ac4b64c5969bd6309d8f8424/natality00.gz
Schema of the CSV and the description of each field can be found HERE. Create a new bucket in Minio, here, we are naming the bucket spark-experiment and upload the downloaded file to that bucket. You can use Minio UI for the task. Or, you can use Minio Client - mc for the same.
# Go to the `data` folder which Minio Server is pointing to
cd ~/data
# Creating a new bucket
mc mb spark-experiment
# Copying the compressed file inside the bucket
mc cp ../natality00.gz spark-experiment
Now, let’s try the following script in Jupyter notebook. You can either create a new cell in the same old notebook or create a new notebook for running the script.
import findspark
findspark.init()
import pyspark
from pyspark.sql.types import *
from pyspark.context import SparkContext
from pyspark.sql.session import SparkSession
sc = SparkContext.getOrCreate()
spark = SparkSession(sc)
df = spark.read.format("csv").option("header", "true").load("s3a://spark-experiment/natality00.gz")
query="SELECT is_male, count(*) as count, AVG(weight_pounds) AS avg_weight FROM natality GROUP BY is_male"
df.createOrReplaceTempView("natality")
df2 = spark.sql(query)
df2.show()
Upon running the script in the notebook, you should get the following output:
+-------+-------+-----------------+
|is_male| count| avg_weight|
+-------+-------+-----------------+
| false| 975147| 7.17758067338709|
| true|1025104|7.439839161360215|
+-------+-------+-----------------+
Visualization with charts and graphs using Pandas Installation Install Pandas using conda. PySpark dataframe requires pandas >= 0.19.2 for executing any of the features by pandas.
# Installing pandas and matplotlib. Make sure inside the created conda virtual environment, when you are running the following command
conda install pandas matplotlib
Reports and Observations
Report 1
Let’s display some charts on the report that we got in the previous example. Let’s create a new cell on the same notebook rather than integrating the following snippet in the above code, to reduce the time to plot multiple charts on the same report.
df3 = df2.toPandas()
df3.plot(x='is_male', y='count', kind='bar')
df3.plot(x='is_male', y='avg_weight', kind='bar')
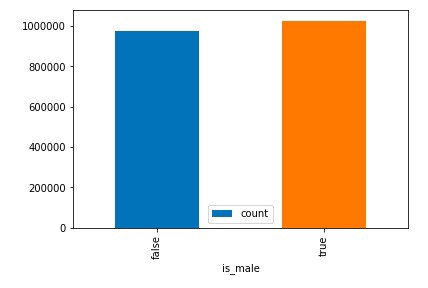
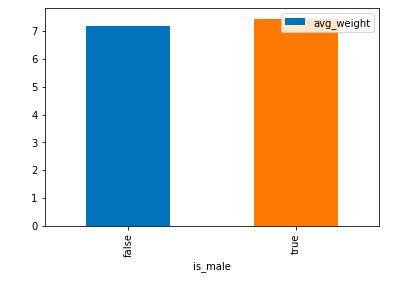 Chart Graph of Is_Male Boolean VS Count and Average weight
Chart Graph of Is_Male Boolean VS Count and Average weight
Observation: From the generated chart, we can observe that gender of the child doesn’t have any significant role neither in the average weight of the child nor wide difference can be seen in a total count of the two gender divisions.
Report 2
Now, let us try another example. Let’s create a new notebook for this. If you don’t wish to create a new one, you can try on a new cell of the previous notebook.
import findspark
findspark.init()
import pyspark
from pyspark.sql.types import *
from pyspark.context import SparkContext
from pyspark.sql.session import SparkSession
sc = SparkContext.getOrCreate()
spark = SparkSession(sc)
df = spark.read.format("csv").option("header", "true").load("s3a://spark-experiment/natality00.gz")
query="SELECT mother_age, count(*) as count, AVG(weight_pounds) AS avg_weight FROM natality GROUP BY mother_age"
df.createOrReplaceTempView("natality")
print("Based on mother_age, total count and average weight is as follow : ")
df2 = spark.sql(query)
df3 = df2.toPandas()
df4= df3.sort_values('mother_age')
print("***DONE***")
After running the program, when it prints DONE. Create a new cell below and run the following snippet:
df4.plot(x='mother_age', y='count')
df4.plot(x='mother_age', y='avg_weight')data
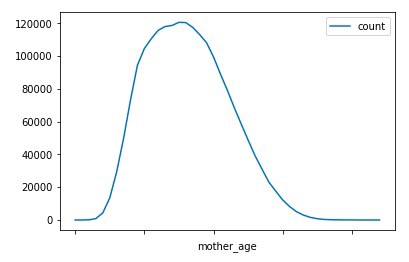
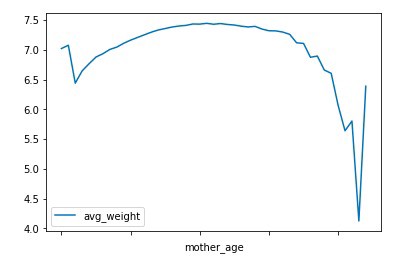 Chart Graph of Mother Age VS Count and Average weight
Chart Graph of Mother Age VS Count and Average weight
Observation : We can observe that most of the mothers are between 20–30 age range when they gave birth. While the average weight of the children shows some decline in case of mothers at a young age, it shows a significant decrease in children’s average weight in case of mothers at old age.
Report 3
This one will be an interesting one. We will plot a chart with a scatter graph.
import findspark
findspark.init()
import pyspark
from pyspark.sql.types import *
from pyspark.context import SparkContext
from pyspark.sql.session import SparkSession
sc = SparkContext.getOrCreate()
spark = SparkSession(sc)
df = spark.read.format("csv").option("header", "true").load("s3a://spark-experiment/natality00.gz")
query="SELECT INT(gestation_weeks), COUNT(*) AS count, AVG(weight_pounds) AS avg_weight FROM natality GROUP BY gestation_weeks"
df.createOrReplaceTempView("natality")
print("Based on gestation_weeks, total count and average weight is as follow : ")
df2 = spark.sql(query)
df3 = df2.toPandas()
df4= df3.sort_values('gestation_weeks')
print("***DONE***")
Like we did before, after DONE is printed. Create a new cell below with the following snippet. Here, we are introducing matplotlib’s axes object(ax), and dataframe.describe().
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
df4.plot(kind="scatter", x="gestation_weeks", y="avg_weight", s=100, c="count", cmap="RdYlGn", ax=ax)
df4.describe()
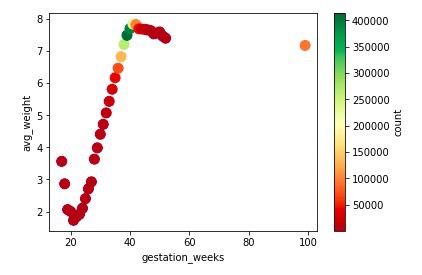
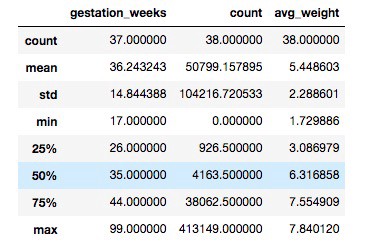 Scatter Graph and Data Frame Description of Gestation week VS Count and Average weight
Scatter Graph and Data Frame Description of Gestation week VS Count and Average weight
Observation: From the scatter graph, it can be seen that the maximum number of mothers'
gestation period was 40 weeks and children born around this period are mostly of more
weight than rest. It can be seen that there are around 100k entries for which gestation_weeks
is 99, which is not possible in reality. So, it can be concluded that 99 is the dummy
value present for those whose gestation period data wasn’t available.
Note: List of possible cmap i.e. colormap can be found here.